Quantum chemical calculations
We study catalytic mechanisms of enzymes using quantum chemical calculations.
- Analysis of catalytic mechanism of C-methyltransferase Fur6
- Analysis of mechanism of stereoselectivity in strigolactone biosynthesis
Analysis of catalytic mechanism of C-methyltransferase Fur6
A methyltransferase Fur6 that functions in the biosynthetic pathway of an antitumor compound, furaquinocin D, transfers a methyl group from S-adenosylmethionine (SAM) to C3 of 1,2,4,5,7-pentahydroxynaphtalene (PHN) (Fig.1). In this study, we constructed a structural model of the ternary complex of Fur6 with SAM and PHN based on the crystal structure of the binary complex between Fur6 and S-adenosyl-l-homocysteine (SAH), which is produce after SAM donates a methyl group. After we confirmed the stability of the structural model using MD simulation, we conducted QM/MM calculations to analyze the catalytic mechanism (Fig. 2). We found that the activation energy was 8.6 kcal/mol, which is consistent with the experimental fact that the reaction proceeds at a room temperature. This study was conducted in collaboration with the Synthetic Biology Laboratory of the University of Tokyo.
Zhao et al., Biochemistry, 63, 806–814 (2024).
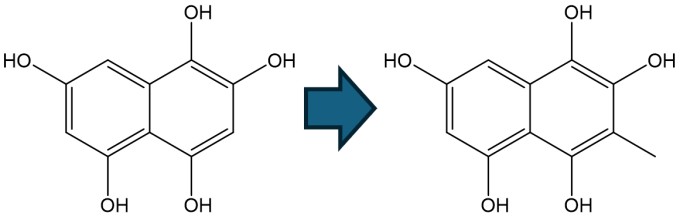
Fig. 1: The structures of the substrate PHN (left) and the product (right).
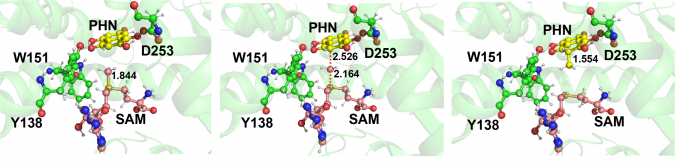
Fig. 2: The structures of the reactant (left), transition state (middle), and the intermediate (right). The relative energies of the transition state and the intermediate with respect to the reactant were 8.6 kcal/mol and −32.9 kcal/mol, respectively. The intermediate has an extra proton on C3, which is removed when the intermediate is ejected to the solvent.
Analysis of mechanism of stereoselectivity in strigolactone biosynthesis
Strigolactones (SLs) are plant hormones with four rings of A, B, C, and D. SLs are produced from a precursor, 18-oxo-carlactonoic acid (18-oxo-CLA) at the final step of their synthesis, where closure of BC-ring occurs (Fig. 3). When this reaction occurs in flask, a mixture of two diastereomers, the strigol and the orobanchol types, is produced at a ratio of 56% and 44%. Prof. Sugimoto’s group at Kobe University identified an enzyme termed SlSRF that catalyzes this reaction but produces only orobanchol-type SLs. Aming at clarifying the mechanism of the stereoselectivity of SlSRF, we conducted a collaborative research with them.
In this study, we first predicted the tertiary structure of SlSRF using AlphaFold and then constructed a structural model of the complex between SlSRF and the substrate. Subsequently, we conducted an MD simulation for the complex. We found that the substrate stably adopted the conformation favorable to producing the orobanchol-type product. In contrast, the substrate adopted the conformation favorable to producing the strigol-type product as well as the conformation favorable to producing the orobanchol-type product during the MD simulation of the precursor alone in solution (Fig. 4). Because this result is consistent with that of the experiment in flask, we concluded that the stereochemistry of the product was determined by the conformation of the substrate before the reaction. We next performed QM/MM calculations to analyze the catalytic mechanism of SlSRF. We found that the reaction proceeded through transition state 1, intermediate, and transition state 2. The maximum activation energy with respect to the reactant’s energy was 14.6 kcal/mol, which is consistent with the fact that the reaction proceeds at a room temperature (Fig. 5).
Homma et al., Proc. Natl. Acad. Sci. USA, 121, e2313683121 (2024).

Fig. 3: The reaction producing SLs. The reaction occurs by protonation of the precursor, 18-oxo-CLA.
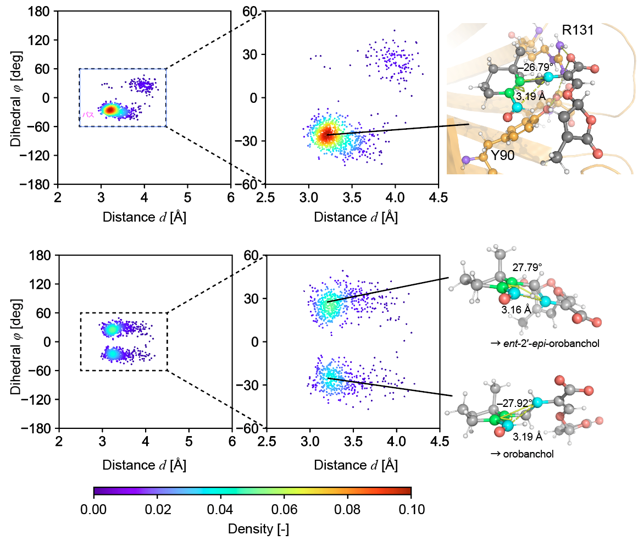
Fig. 4: Conformation distributions of the precursor, 18-oxo-CLA during the MD simulations for the complex (upper) and for the precursor alone in solution (lower). The conformation favorable to producing the orobanchol-type product was dominant in the MD simulation for the complex, whereas two conformations were observed in the MD simulation for the precursor alone in solution.
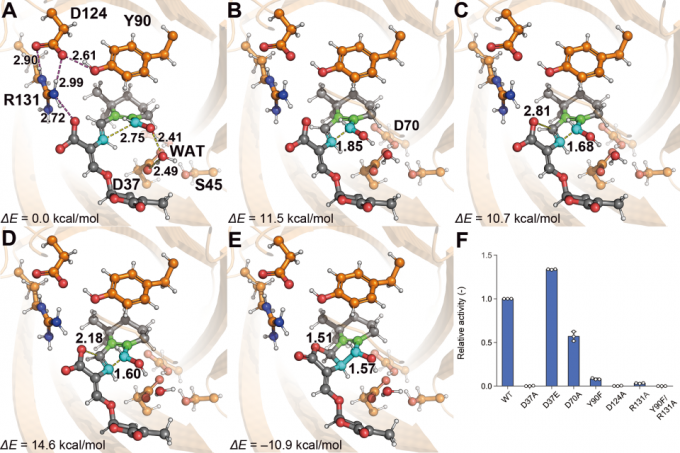
Fig. 5: The structures of (A) reactant, (B) transition state 1, (C) intermediate, (D) transition state 2, and (E) product. (F) Results of the mutation experiment that confirmed the validity of the structural model of the complex.
